USER MANUAL
Table of Contents
1. About RTKs
Signal transduction is an essential process for the survival of all organisms. It regulates several biological functions including cellular proliferation, apoptosis and maintenance of homeostasis. In metazoa, a protein family with a significant role in signal transduction includes protein tyrosine kinases (PTKs). A distinguishable group of PTKs is formed by transmembrane receptor tyrosine kinases (RTKs). Their extracellular N-terminus constitutes the binding site for hormones and growth factors, while their intracellular C-terminus catalyzes the phosphorylation of tyrosine residues. RTKs have been implicated in severe diseases such as cancer, neurodegenerative and cardiovascular diseases (Mark Lemmon and Joseph Schlessinger, 2010).
| ⦗Back to Top⦘ |
2. How RTK-PRED works
RTK-PRED is an algorithm that combines profile Hidden Makov Models (pHMM) with a transmembrane topology prediction algorithm, PHOBIUS (http://phobius.sbc.su.se/). The main feature of this algorithm is the detection of Protein Tyrosine Kinase Domains using a specialized pHMM. Afterwards, PHOBIUS predicts the topology of the sequence and the existence and position of extracellular domains. Finally, the latter are used for the functional annotation/classification of the receptor.
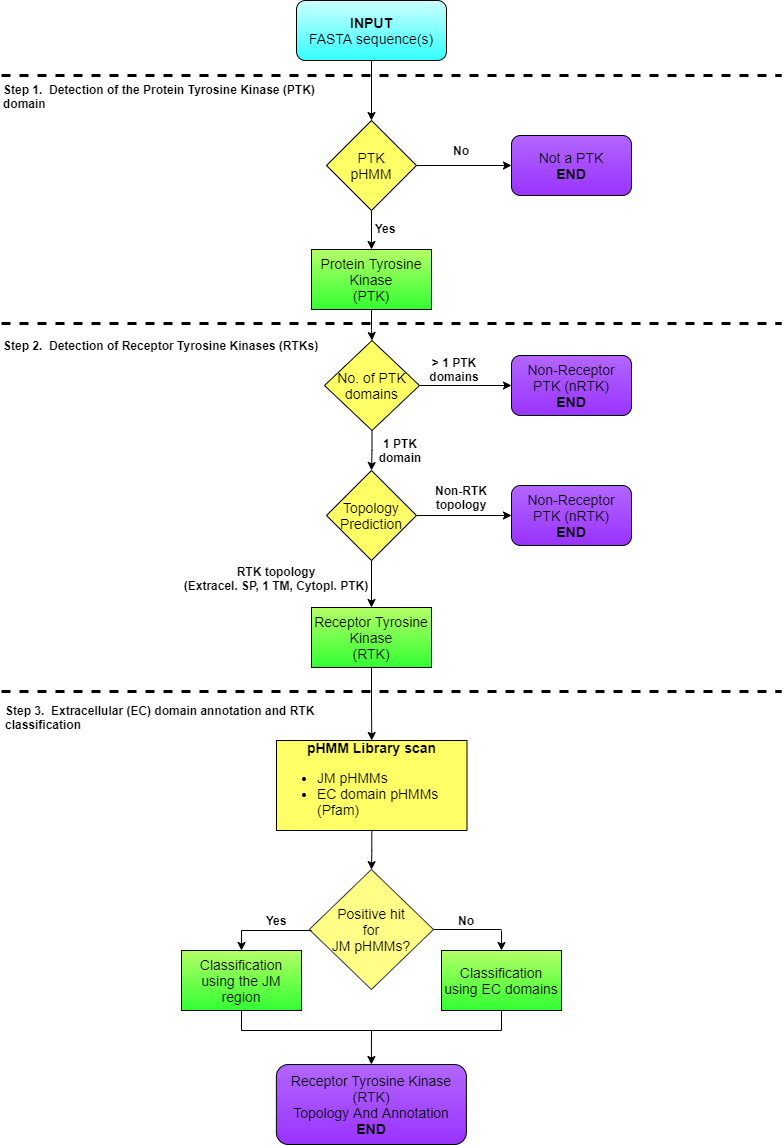
Figure 1. RTK-PRED's flowchart
| ⦗Back to Top⦘ |
3. Profile Hidden Markov Models (pHMMs)
For the training and validation of the specific Tyrosine Kinase Domain pHMM, we used HMMER v3.0 (http://hmmer.org/) alongside with HMM-ModE, a computational method that generates family specific pHMMs using negative training sequences. Below, there are the sequences we used to train the pHMM which RTK-PRED is using.
Positive training set for the training of the Tyrosine Kinase Domain specific pHMM.
Negative training set for the training of the Tyrosine Kinase Domain specific pHMM.
To classify RTKs into subfamilies (Figure 2), RTK-PRED uses several pHMMs which are corresponding to different domains in the extracellular region of each RTK subfamily. Those pHMMs were downloaded from PFAM (https://pfam.xfam.org/) and are presented in the following able.
| PFAM ID | PFAM Accession |
|---|---|
| Receptor_L_domain | PF01030 |
| Cadherin | PF00028 |
| Furin-like | PF00757 |
| ig | PF00047 |
| fn3 | PF00041 |
| Sema | PF01303 |
| TIG | PF01833 |
| LRR_8 | PF13855 |
| F5_F8_type_C | PF00754 |
| Fz | PF01534 |
| Kringle | PF00051 |
| Ldl_recept_a | PF00057 |
| MAM | PF00629 |
| ⦗Back to Top⦘ |
4. Using RTK-PRED
RTK-PRED is publicly available through: http://bioinformatics.biol.uoa.gr/RTK-PRED/.
4.1 User Input
The RTK-PRED input interface is shown in Figure 2.
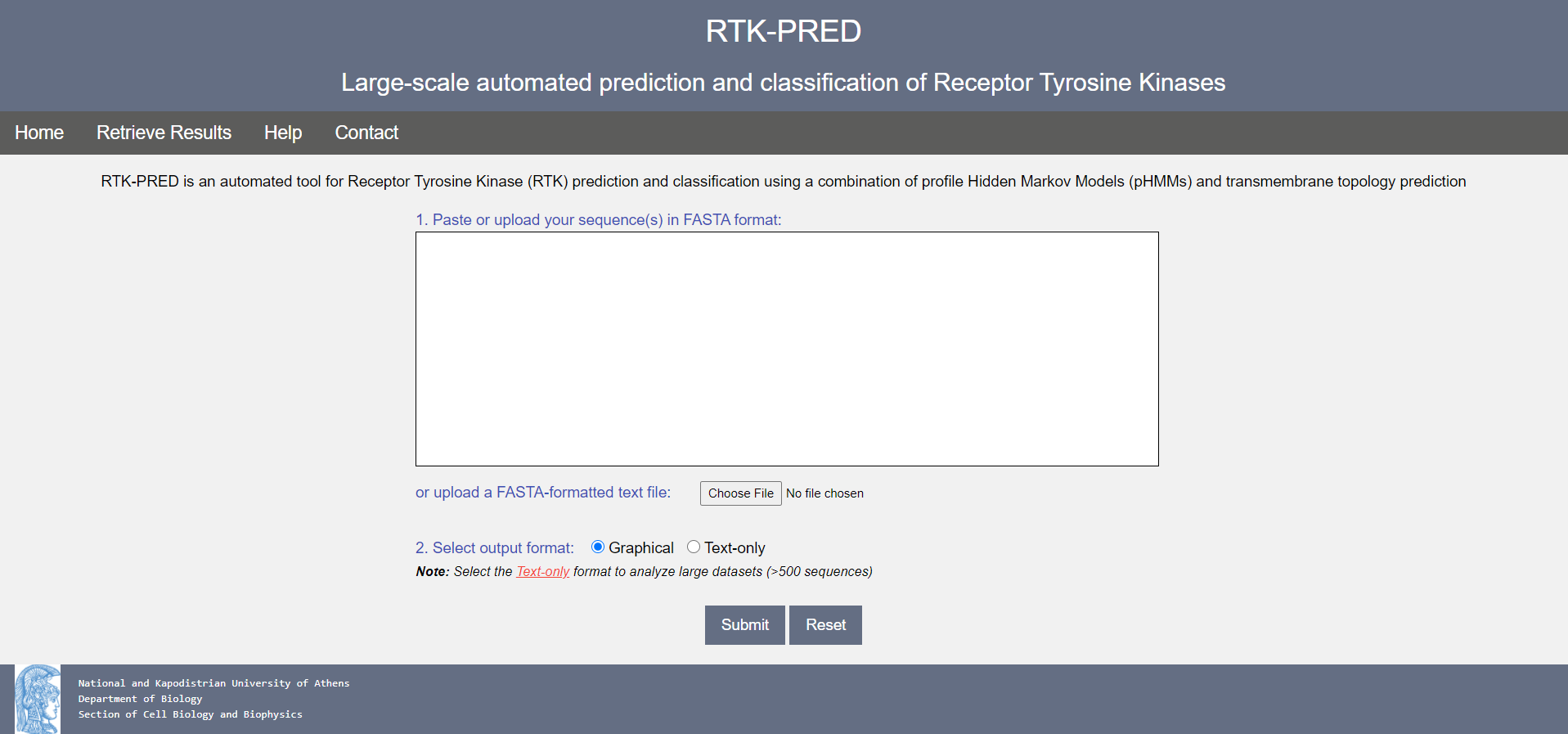
Figure 2. The RTK-PRED user input page
Instructions to use:
- Paste your sequence(s) in FASTA format in the text submission form or upload a FASTA-formatted sequence file (note: file size must not exceed 40 MBs).
- Select the output format. Two options are offered:
- Graphical: The output will be shown in graphical format. Recommended for small-to-medium sequence datasets (up to 500 sequences). This is the default option
- Text-only: A simple, parsable text output will be returned. This option is recommended when dealing with large datasets (>500 sequences) and/or when RTK-PRED results need to be parsed with text processing scripts.
- Press the Submit button to submit your data or the Reset button to clear the form.
| ⦗Back to Top⦘ |
4.2 Results
Upon submission, each RTK-PRED receives a unique numerical identifier (job ID). This ID can be used to retrieve previous run results from the Retrieve Results page.
Depending on the size of your input and the server workload, RTK-PRED jobs will run from a few seconds to a couple of minutes. After the run is completed, you will redirected to the results. Below is an example of the result page RTK-PRED will generate if the input is an RTK (sequence number 1), an nRTK (sequence number 2) and a not tyrosine kinase protein (sequence number 3):
4.2.1 Graphical Format
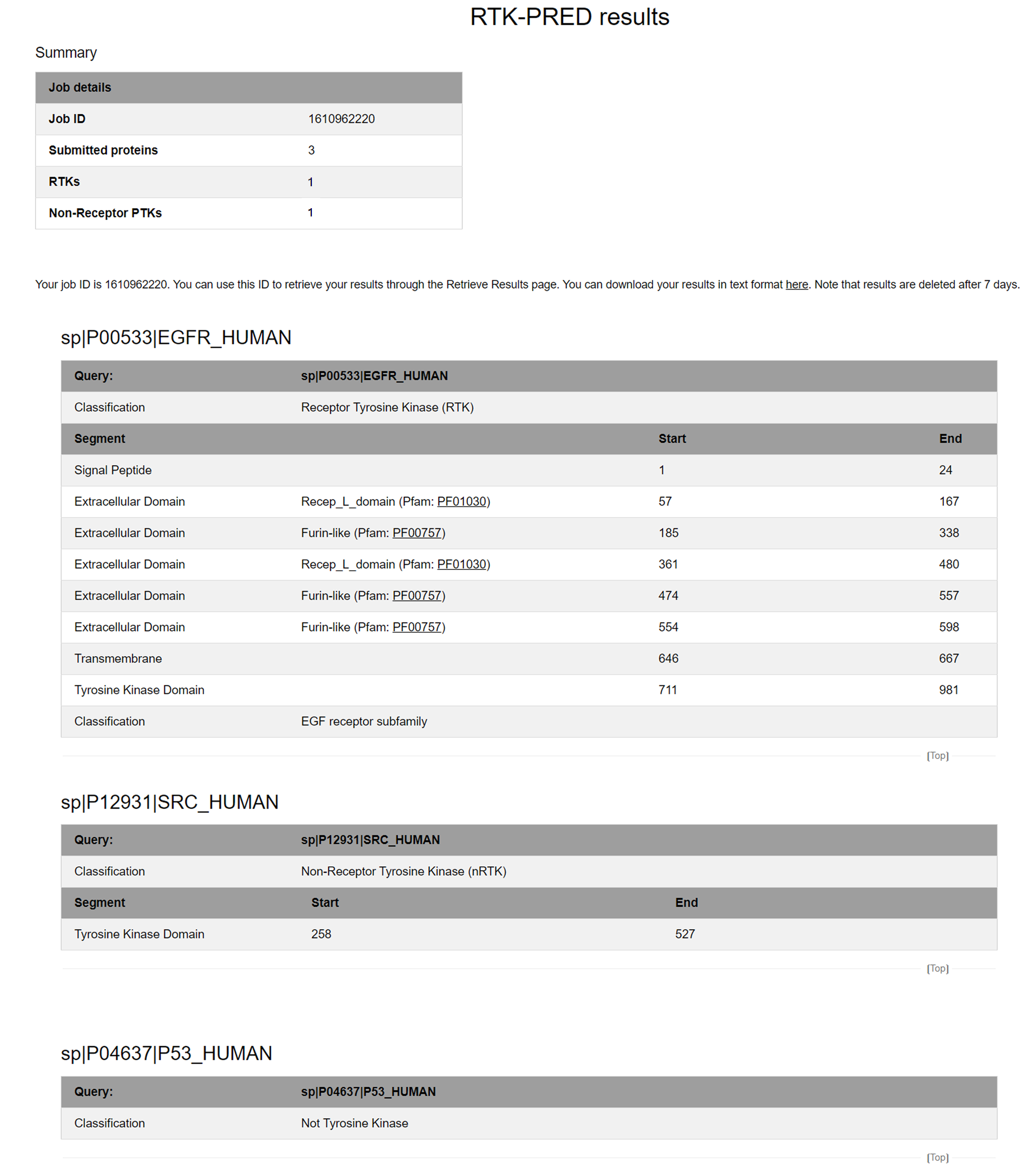
Figure 3. The RTK-PRED results page
4.2.2 Text Format
#Your job ID is 1610962220.
#You can use this ID to retrieve your results through the Retrieve Results page.
#Note that results are deleted after 7 days.
>> Query: sp|P00533|EGFR_HUMAN
Classification: Receptor Tyrosine Kinase (RTK)
SIGNALP: From:1 To:24
EC Domains (PF01030): From:57 To:167 Recep_L_domain Score: 105.2
EC Domains (PF00757): From:185 To:338 Furin-like Score: 101.3
EC Domains (PF01030): From:361 To:480 Recep_L_domain Score: 96.5
EC Domains (PF00757): From:474 To:557 Furin-like Score: 23.8
EC Domains (PF00757): From:554 To:598 Furin-like Score: 9.5
TRANSMEM: From:646 To:667
Kinase Domain: From:711 To:981 Score: 301.3
RTK subfamily: Type 1 (EGF receptor subfamily)
//
>> Query: sp|P12931|SRC_HUMAN
Classification: Non-Receptor Tyrosine Kinase (nRTK)
Kinase Domain: From:258 To:527 Score: 332.5
//
>> Query: sp|P04637|P53_HUMAN
Classification: Not Tyrosine Kinase
//
If a sequence is classified as an RTK (sequence number 1), in each line you can see:
1. Sequence Name
2. Classification between RTKs, nRTKs, Not Tyrosine Kinase
3. Tyrosine Kinase Domain Prediction
4. Tranmembrane Region Prediction
5. Extracellular Domain Predictions
6. Classification between the 18 mammalian RTKs' subfamilies
Each result starts with double greater-than sign ">>" and ends with double backslash sign "//". Also, the results are terminated earlier
when a non RTK protein is uploaded (sequences number 2 and number 3).
Please note that RTK-PRED results are automatically deleted after seven days.
| ⦗Back to Top⦘ |
4.3 Retrieve Results
The user may also retrieve his results from a past run, using the unique Job ID provided (see previous section) and the Retrieve Results page:
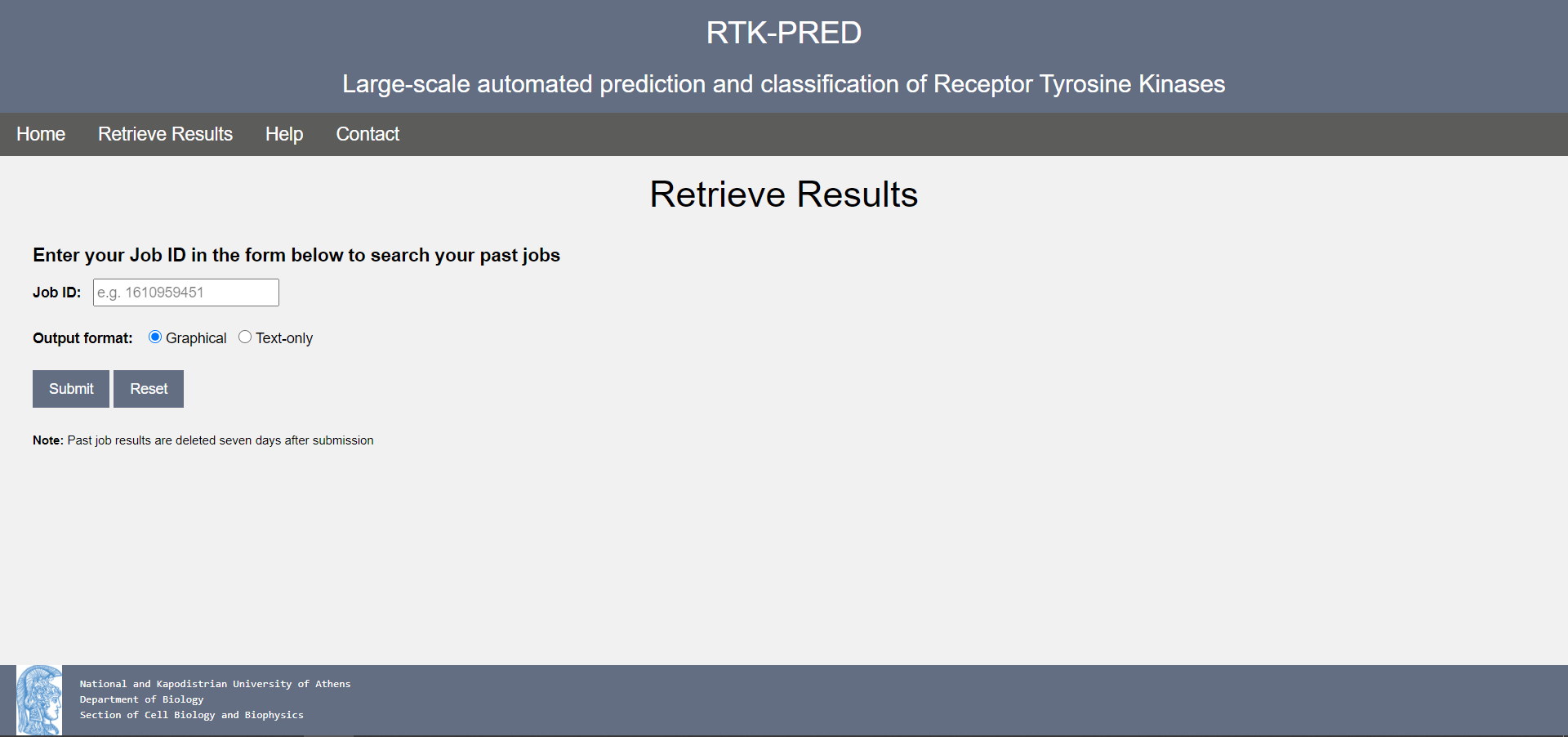
Figure 4. The RTK-PRED Retrieve Results page
To retrieve past results, the user needs to submit his Job ID in the form and choose the output format (graphical or text). If a valid job ID is submitted (and/or the RTK-PRED job was run less than seven days before), the user will be redirected to their results. Otherwise, a helpful error page will be displayed.
| ⦗Back to Top⦘ |
5. Privacy Policy
RTK-PRED collects some user data to successfully provide its services. For more information, please consult the service's privacy notice.
